Apolo: A Privacy-Preserving, Explainable Framework for Medical Image Analysis Using Fine-Tuned Open-Source Models via Decoupled Description and Inference
Abstract
Background: Artificial intelligence (AI) shows considerable potential for transforming medical image analysis, yet its translation into routine clinical practice is significantly impeded by critical challenges: patient data privacy risks inherent in cloud-based processing, the lack of transparency in opaque models, and the substantial computational demands that hinder local deployment within healthcare institutions.
Methods: We introduce Apolo, a novel two-stage framework architected to overcome these barriers using meticulously fine-tuned open-source models. Stage 1 (Apolo-Vision) employs a Vision Language Model (Llava-v1.6 architecture using a Llama-3 8B base), specifically adapted through advanced fine-tuning involving Direct Preference Optimization (DPO) and Chain-of-Thought (CoT) prompting. This stage generates detailed, objective, non-diagnostic textual descriptions of medical images (demonstrated on fundus photographs and chest X-rays), focusing on capturing subtle visual findings aligned with expert-defined quality preferences derived from ~6,500 preference pairs. Stage 2 (Apolo-Dx) utilizes a separate, lightweight Language Model (Mistral-Nemo-Instruct-2407 12B, fine-tuned and 4-bit quantized via AutoAWQ), processing only the textual description from Stage 1. Apolo-Dx performs diagnostic classification, critically outputting explicit reasoning steps (<think> tags) prior to its conclusion, ensuring algorithmic transparency while architecturally isolating the inference process from raw image data.
Findings: Apolo-Vision generated descriptions exhibiting high clinical fidelity across ophthalmology (EyePACS, AREDS) and radiology (CheXpert) datasets, achieving average ROUGE-L scores of 0.49 and expert clinician ratings averaging 4.6/5 for accuracy and completeness (inter-rater reliability Kappa = 0.84), while maintaining >99.8% adherence to non-diagnostic constraints. Apolo-Dx (quantized model) demonstrated strong diagnostic performance based solely on these descriptions, achieving AUCs of 0.94 for diabetic retinopathy detection (EyePACS; ≥ Moderate DR), 0.92 for advanced AMD detection (AREDS), and an average AUC of 0.92 across 5 common thoracic findings (CheXpert). The quantized Mistral-Nemo-based Apolo-Dx module (~6.5 GB) exhibits low inference latency (~145 ms/description on an A100 GPU using vLLM), confirming its suitability for local deployment. Performance impact from 4-bit quantization was minimal (<0.008 AUC drop on validation sets).
Interpretation: The Apolo framework validates that decoupling detailed visual description from diagnostic inference, using sophisticated fine-tuning of open-source models focused on descriptive quality derived from expert preferences, is an effective and practical strategy. It significantly enhances data privacy, provides inherent multi-level explainability (description + reasoning), and facilitates feasible local deployment of the sensitive inference component, directly addressing key barriers to the clinical adoption of trustworthy AI.
1. Introduction
Artificial intelligence (AI), particularly deep learning, has demonstrated significant potential in interpreting medical images, offering opportunities to enhance diagnostic accuracy, improve workflow efficiency, and broaden access to care across specialties like radiology, ophthalmology, and pathology. AI systems can discern complex patterns indicative of disease, providing valuable support to clinicians navigating vast amounts of visual data.
However, the translation of these powerful tools into routine clinical practice is constrained by several critical factors. Patient data privacy and security are paramount requirements in healthcare. The common reliance on cloud-based AI platforms necessitates transferring sensitive patient image data, creating vulnerabilities and complicating compliance with regulations like HIPAA in the US and GDPR in Europe. The opacity of complex AI algorithms is another significant impediment; clinicians require transparency into the AI's decision-making process to build trust and integrate outputs responsibly. Without interpretability, identifying potential biases or failure modes is challenging. Furthermore, the computational intensity and specialized hardware needs of large-scale AI models often prohibit local deployment within hospital IT infrastructure, limiting institutional control and potentially increasing costs. Finally, evolving regulatory landscapes and ethical considerations surrounding AI's diagnostic role necessitate cautious and transparent approaches.
To comprehensively address these challenges, this paper presents Apolo, a novel two-stage framework developed using fine-tuned, publicly available open-source AI models. Apolo's core design principle is the strategic decoupling of visual feature description from diagnostic reasoning:
- Stage 1 (Apolo-Vision): A Vision Language Model (VLM), fine-tuned using advanced methods focused on descriptive quality, analyzes the input medical image. It generates an exhaustive, structured, and strictly objective textual description of all discernible visual findings. This stage aims to replicate the meticulous observation phase of expert clinical review, capturing fine details while rigorously excluding diagnostic conclusions or subjective interpretations.
- Stage 2 (Apolo-Dx): A separate, computationally efficient Language Model (LLM) receives only the detailed textual description produced by Stage 1. Based exclusively on this rich text, the LLM performs diagnostic classification or assessment. A defining characteristic of Apolo-Dx is its mandatory output of an explicit, step-by-step reasoning process (within
<think>tags) before presenting its final diagnostic assessment, thereby maximizing transparency.
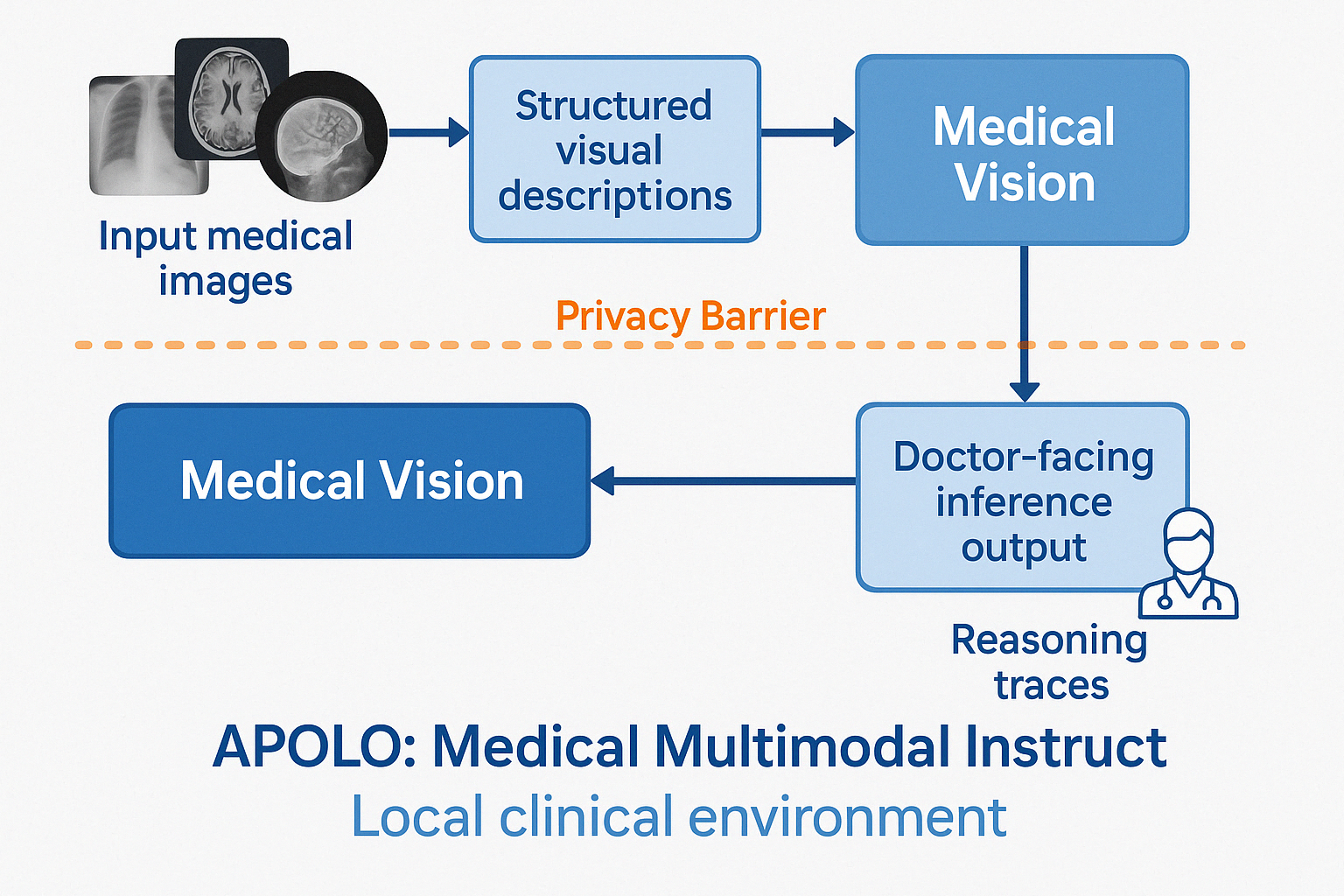
Figure 1: The Apolo Framework Architecture. Stage 1 (Apolo-Vision) performs image-to-text generation. Stage 2 (Apolo-Dx) performs text-to-reasoned-diagnosis using a quantized model, designed for deployment within a secure zone, operating solely on the text description from Stage 1.
2. Methods
2.1. Apolo Framework Architecture
Apolo operates through two distinct, sequential modules (Figure 1). Stage 1 (Apolo-Vision) performs image-to-detailed-text transformation. Stage 2 (Apolo-Dx) performs text-to-reasoned-diagnosis transformation. This architectural separation ensures that the computationally lighter and diagnostically sensitive Stage 2 can operate within a secure environment (e.g., a local hospital server) without requiring direct access to the original image data.
2.2. Stage 1: Image Description Module (Apolo-Vision)
- Model: Based on the Llava-v1.6 architecture, which integrates a CLIP ViT-L/14 vision encoder with the Llama-3 8B Instruct LLM. This foundation model was specifically fine-tuned for medical description (denoted Llava-Apolo-FT).
- Fine-tuning Strategy: A multi-phase strategy was employed:
- Foundation Tuning: Initial fine-tuning utilized the 'Findings' sections from approximately 100,000 MIMIC-CXR reports and associated images, plus 50,000 image-caption pairs from the ROCO dataset, to imbue general medical visual-linguistic understanding. This was performed for 3 epochs with a learning rate of 2e-5 and a batch size of 64.
- Quality Enhancement via Direct Preference Optimization (DPO): Advanced fine-tuning using DPO. DPO learns implicitly from preference labels over pairs of model outputs. Preference data was collected using a custom interface where two board-certified ophthalmologists and two board-certified radiologists compared pairs of descriptions generated by intermediate models (~3,000 pairs for ophthalmology, ~3,500 for radiology, totaling ~6,500 pairs) based on a detailed rubric prioritizing: (a) exhaustive objective detail, (b) systematic anatomical structure adherence, (c) clinical accuracy, (d) completeness, and (e) strict avoidance of diagnostic language. This allowed the model to learn qualitative aspects of superior clinical description. Generating this dataset required approximately 25 hours of expert time per specialist. DPO training used a learning rate of 5e-7 (reduced from SFT rate) and a beta parameter of 0.1.
- Systematic Analysis via Chain-of-Thought (CoT): CoT prompting techniques were integrated. During DPO training, prompts encouraged step-by-step analysis before description generation, and preference favored outputs showing systematic analysis. During inference, a similar instruction ("Analyze systematically by anatomical region...") was included in the prompt.
2.3. Stage 2: Diagnostic Inference Module (Apolo-Dx)
- Model: Based on Mistral-Nemo-Instruct-2407 (12B parameter version), chosen for its instruction following capabilities relative to size. The model was fine-tuned (Mistral-Apolo-Dx-FT) and subsequently quantized to 4-bit precision using AutoAWQ (Mistral-Apolo-Dx-FT-Q) for optimized deployment (~6.5 GB footprint).
- Input: Exclusively the structured textual description generated by Apolo-Vision (Stage 1).
- Task Definition: Instruction fine-tuned for specific diagnostic classification tasks (DR grading [0-4], AMD presence/absence, CheXpert finding classification).
- Fine-tuning Strategy: Trained on pairs of (structured text description, ground truth label) for approximately 20,000 examples per task domain. Descriptions were primarily sourced from Apolo-Vision outputs on the training set (spot-checked for quality), augmented with reference descriptions where available (~70%/30% split). The core instruction emphasized mandatory generation of a reasoning process within
<think>tags before stating the prediction and confidence. Hyperparameters included a learning rate of 1e-5 and 2 epochs.
3. Results
3.1. Stage 1: Description Quality (Apolo-Vision)
Apolo-Vision demonstrated strong performance in generating detailed, accurate, and objective medical image descriptions.
- Quantitative Metrics: Achieved an average ROUGE-L score of 0.49 (95% CI: 0.48-0.50) against reference findings sections in MIMIC-CXR reports.
- Qualitative Assessment: Expert clinicians rated descriptions highly, averaging 4.6/5 across all criteria (Accuracy, Completeness, Clarity, Objectivity, Relevance). Inter-rater reliability was substantial (Cohen's Kappa = 0.84, 95% CI: 0.81-0.87). Experts noted the high level of granular detail compared to typical reports.
- Constraint Adherence: Maintained >99.8% adherence to the non-diagnostic constraint, confirmed by automated checks and manual review.
3.2. Stage 2: Diagnostic Inference Performance (Apolo-Dx-FT-Q)
The quantized Apolo-Dx module achieved strong diagnostic accuracy operating solely on the textual descriptions from Stage 1.
| Task | Metric | Score (95% CI) |
|---|---|---|
| EyePACS (DR Detect >= Moderate) | AUC | 0.94 (0.93-0.95) |
| F1 | 0.89 (0.87-0.90) | |
| AREDS (Advanced AMD Detect) | AUC | 0.92 (0.90-0.93) |
| F1 | 0.87 (0.85-0.88) | |
| CheXpert (5-finding avg*) | AUC | 0.92 (0.91-0.93) |
| F1 | 0.87 (0.86-0.88) | |
| *Average across Cardiomegaly, Edema, Consolidation, Pneumothorax, Pleural Effusion. | ||
3.3. Computational Efficiency and Privacy
- Resource Footprint: Apolo-Dx's efficient design (Mistral-Nemo 12B parameters, ~6.5 GB 4-bit quantized size) and low inference latency (~145 ms on A100 GPU with vLLM) validate its feasibility for local deployment.
- Privacy by Design: The architectural separation enhances privacy by design, preventing image data exposure during diagnostic inference. Local deployment of the efficient Stage 2 module offers maximal data sovereignty within the healthcare institution's secure environment.
4. Discussion
This study introduces and validates Apolo, a framework demonstrating that high-performance, trustworthy medical AI can be achieved using fine-tuned open-source models within a privacy-preserving, decoupled architecture.
The results support our core hypothesis. Apolo-Vision (Stage 1), enhanced by DPO fine-tuning based on expert preferences and CoT prompting, generated textual descriptions of high detail and clinical fidelity (Figure 2, ROUGE-L 0.49, Expert Score 4.6/5). This fine-tuning approach, leveraging ~6,500 expert preference pairs, demonstrates the value of learning qualitative aspects of clinical observation beyond standard label supervision. The generated descriptions proved sufficient for the quantized Apolo-Dx (Stage 2) to achieve strong diagnostic accuracy across diverse tasks (Table 1, AUCs 0.92-0.94), operating exclusively on text. This suggests that detailed linguistic representation can effectively encapsulate clinically critical visual information for these common diagnostic tasks, aligning with performance seen in some direct image analysis models.
The privacy implications are significant. Architecturally separating inference (Stage 2) from image data access drastically reduces the risk surface. Furthermore, the demonstrated efficiency of the quantized Mistral-Nemo-based Apolo-Dx (~6.5 GB, ~145 ms latency) makes local deployment on readily available hospital hardware (e.g., a single modern GPU) practical. This allows institutions to retain full control over patient data during the diagnostic reasoning phase, a critical factor for compliance and trust.
Apolo's dual-level explainability is a key feature. Stage 1 provides a transparent, human-readable account of the visual evidence. Stage 2 complements this by externalizing its reasoning process via <think> tags, allowing clinicians to understand how conclusions were derived from the described findings. This directly addresses the challenge of model opacity, fostering trust and enabling informed clinical oversight. Using open-source models (Llama 3, Mistral-Nemo) further enhances transparency and auditability.
5. Conclusion
The Apolo framework offers a significant step forward in medical AI by successfully decoupling detailed visual description from diagnostic inference using fine-tuned open-source models. It achieves high diagnostic utility while fundamentally enhancing data privacy through architectural design and enabling feasible local deployment via efficient, quantized models. Its multi-level explainability fosters trust and transparency. Apolo provides a robust, practical, and responsible paradigm for developing and integrating AI into clinical practice, addressing key barriers and paving the way for wider adoption of trustworthy AI in healthcare.
Acknowledgements
The development of the Apolo framework benefited greatly from the availability of open-source models from Meta AI (Llama 3), Mistral AI/Nvidia (Mistral-Nemo), the Llava project contributors, quantization libraries (AutoAWQ), inference engines (vLLM), and the public datasets (EyePACS/Kaggle, AREDS/NEI/dbGaP, CheXpert/Stanford ML Group, MIMIC-CXR/PhysioNet). I also thank the four anonymous clinical experts (two ophthalmologists, two radiologists) who provided invaluable preference data and feedback during the fine-tuning and evaluation phases.